No abstract is available for this article.
Oral Peptide Nano‐Formulations for Breast Cancer: An Enhancer‐Centric, Regulatory‐Ready Path to Clinical Translation

Strategic shift to oral delivery: Oral peptide nano-formulations address the critical need for non-invasive, chronic administration. Enhancer-centric design: The review advocates for using permeation enhancers with established regulatory precedence. Focus on reproducibility: Viability in oncology is defined by controlled pharmacokinetic variability and reproducible exposure. Minimalist architecture: Lipid-based nano-platforms that facilitate peptide–enhancer co-localization over complex, novel structures. Translational priorities: Development pathways must prioritize CMC robustness and scale-up feasibility, and for successful commercialization.
ABSTRACT
Breast cancer is increasingly managed as a long-term condition, intensifying the need for therapies that are effective, tolerable, and suitable for chronic administration. Peptide-based therapeutics offer high molecular specificity and favorable safety profiles; however, their clinical utility remains largely limited by reliance on parenteral delivery. Oral administration is strategically attractive for improving patient adherence, quality of life, and healthcare efficiency, yet peptide drugs face formidable gastrointestinal barriers, including enzymatic degradation, restricted epithelial permeability, and substantial pharmacokinetic variability. These challenges may be further exacerbated by treatment-related gastrointestinal dysfunction in oncology populations. This review critically evaluates oral peptide nano-formulations that incorporate molecular permeation enhancers with established regulatory precedence as a pragmatic approach to achieving clinically meaningful oral absorption in breast cancer. Rather than emphasizing formulation novelty, the article adopts a translational, industry-oriented perspective that prioritizes regulatory alignment, chronic-use safety, manufacturability, and CMC robustness. Mechanistic and translational aspects of key enhancer classes—including medium-chain fatty acids, bile-derived agents, and surfactant-based enhancers—are examined, with particular attention to epithelial reversibility, dose–response nonlinearity, gastrointestinal tolerability, and exposure consistency. Evidence supports an enhancer-centric paradigm which primarily serves to protect peptides and co-localize peptide–enhancer exposure at the intestinal absorption site.
Peptide Mapping Using Multienzyme Digestion Strategies Integrated with LC‐HRMS Workflow: A Case Study

Exenatide and H-GLP-1 belong to the GLP-1 agonist class of potential therapeutic agents. A robust multienzyme peptide-mapping workflow combined with LC-HRMS/MS enables high-resolution sequence coverage and monitoring of post-translational modifications, providing confident structural verification and comprehensive quality evaluation.
ABSTRACT
Peptide therapeutics are becoming an increasingly important class of pharmaceuticals due to their attractive therapeutic properties. However, their development remains challenging because of inherent structural complexity, requiring advanced analytical techniques. Peptide mapping is a key method for comprehensive identification of a peptide’s primary structure, and FDA and EMA recommend its use to demonstrate structural “sameness.” In this study, we describe a multienzyme peptide-mapping workflow designed to improve sequence coverage and structural verification. The protocol integrates multienzymatic digestion with complementary specificity, enabling generation of alternative peptide maps. Waters XBridge Peptide BEH C18 column (300 Å, 2.5 μm, 4.6 × 150 mm) with gradient separation program was used for separation. Further, LC-HRMS (Orbitrap) was employed to obtain high-resolution MS/MS data, achieving precise peptide fragment identification with mass accuracy within < 5 ppm. Two antidiabetic peptides, exenatide and H-GLP-1, were selected as model systems. Full sequence coverage of both peptides was achieved by combining peptide maps produced with trypsin, Glu-C, and chymotrypsin. Application of the workflow to a GLP-1 analog provided > 95% sequence coverage and accurate intact-mass confirmation. Overall, the described method offers a reliable and stepwise approach for peptide-mapping-based structural verification, enabling confident assessment of the primary structure of exenatide and H-GLP-1.
Conseil d’Administration
The GFPP is administered by a scientific committee composed of members elected at the general assembly held at each congress, as well as the delegate from France to the scientific council of the European Peptide Society (EPS), which is elected by the French members of the EPS. GFPP board members 2025-2027 Maud Larregola President …
Continue reading “Conseil d’Administration”
L’article Conseil d’Administration est apparu en premier sur Groupe Français des Peptides et des Protéines.
GFPP Board
The GFPP is administered by a scientific committee composed of members elected at the general assembly held at each congress, as well as the delegate from France to the scientific council of the European Peptide Society (EPS), which is elected by the French members of the EPS.
GFPP board members 2025-2027
 |
|
|
François Hallé
Vice-President
Assistant Professor in Medicinal Chemistry
Equipe COSSBA – UMR 5246 ICBMS Université Claude Bernard Lyon 1
ISPB – Faculté de Pharmacie
Bat. Rockefeller, 4ème étage, couloir A
8, avenue Rockefeller
F-69373 LYON cedex 8
c
|
 |
| Christophe André Social networks Program Leader Innovation in Innovation Department Polypeptide Bioparc 3, 850 Bd Sebastien Brant 67400 Illkirch, France |
 |
|
Prisca Boisguérin
Newsletter and broadcast list
Director of research at CNRS
|
 |
| Charlène Gadais Secretary Maîtresse de Conférences Institut des Sciences Chimiques de Rennes UMR CNRS 6226 Université de Rennes Faculté de Pharmacie – Campus de Villejean – Bât.6 R+4 2 av. du Pr. Léon Bernard 35043 Rennes Cedex website |
 |
|
Céline Landon
Newsletter and broadcast list
|
 |
|
Baptiste Legrand
Newsletter and broadcast list
Institut des Biomolécules Max Mousseron (IBMM)
UMR CNRS 5247, Université de Montpellier (UM) Pôle Chimie Balard Recherche 1919, route de Mende 34293 MONTPELLIER cedex 5 Phone: +33 (0)4 48 79 21 85
|
 |
|
Olivier Lequin
Treasurer
Laboratoire Chimie Physique et Chimie du Vivant (CPCV) |
 |
| Oleg Melnyk French national representative at the European Peptide Society (EPS) Webmaster Director of research at CNRS Center for Infection and Immunity Pasteur Institute of Lille 1 rue du Pr Calmette 59021 Lille, France Phone : +33 (0)3 20 87 12 14 web site |
 |
| Roba Moumné Webteam Maîtresse de Conférences, HDR Sorbonne Université UMR 8228, Laboratoire CPCV Tour 23-33 – 5ème étage – Porte 506 4 place Jussieu 75005 Paris Cedex 5 |
 |
 |
|
| Loïc Stephan Webteam Researcher at CNRS Laboratoire de Chimie-Physique Macromoléculaire LCPM UMR CNRS 7375 1 rue Grandville 54000 Nancy Phone : +33 (0)3 72 74 37 07 website |
 |
|
Nicolo Tonali
Research Fellow at CEA Saclay |
 |
Improvement of Analysis and Transferability in Peptide Purification: From HPLC to FPLC and Back Again

A predictive workflow enables accurate transfer from analytical HPLC to preparative FPLC for peptide purification. Systematic evaluation of key chromatographic parameters identified particle size, flow optimization and modifier choice as critical for selectivity and resolution. A correction model minimized elution transfer errors from −17% to less than −5%, achieving first-pass purifications with purities higher than > 90%.
ABSTRACT
Optimal purification, high purity and robust purity control for synthetic peptides are critical, as even minor impurities can alter biological activity, distort analytical results and compromise downstream applications. Reliable translation of analytical HPLC results to preparative FPLC conditions remains challenging due to system-specific differences in column geometry, gradient formation and mobile-phase chemistry. This study addresses these limitations through a systematic evaluation of chromatographic parameters influencing peptide selectivity, resolution and transferability. Using a defined peptide impurity library, the effects of gradient steepness, flow rate, temperature and mobile-phase modifier were quantified, revealing that flow-rate optimization and modifier choice have the greatest impact on separation quality. A correction equation was developed to compensate for system-dependent deviations, reducing transfer errors in elution percentage from approximately 17% to less than 5%. The optimized workflow enabled initial preparative purifications with purities above 90% and yields exceeding 30%. Additionally, substitution of trifluoroacetic acid with formic acid was explored as a greener modifier, providing selective improvements in separation performance. The approach establishes a practical and sustainable workflow for the transfer of HPLC-to-FPLC methods for peptide purification.
Covalent Peptide‐Based N‐Myc/Aurora‐A Inhibitors Bearing Sulfonyl Fluoride Warheads

N-Myc-derived peptidomimetics bearing aryl sulfonyl fluoride warheads were shown to act as effective N-Myc/Aurora-A PPI inhibitors, selectively labelling Aurora-A in a recognition-directed manner.
ABSTRACT
Orthosteric inhibition of the N-Myc/Aurora-A protein–protein interaction (PPI) represents a potential mechanism by which degradation of N-Myc can be induced, given its interaction with Aurora-A competes with the factors that tag it for proteasomal degradation. As such, this would constitute an approach for the development of drugs to treat neuroblastoma, a childhood cancer that depends upon N-Myc. Reactive electrophiles have proven useful in the context of targeted covalent inhibitors, and in this work, we sought to improve the potency of a previously identified N-Myc-derived peptide by introducing a sulfonyl fluoride warhead. We successfully demonstrated selective labelling of Aurora-A using the resultant peptidomimetics and established this labelling as recognition-directed, providing valuable insight for further future development of N-Myc peptidomimetics and further broadening the use of aryl sulfonyl fluoride warheads in the context of peptidomimetic PPI inhibitors.
Bismuth Bicycles

Discovery, evolution and potential future applications of bismuth bicycle molecules—a novel class of therapeutics with unique properties.
ABSTRACT
Bicyclic peptides are emerging as next generation therapeutics by combining the affinity and specificity of antibodies with the synthetic convenience of small molecules. Phage-encoded libraries of bicyclic peptides enable the discovery of high-affinity molecules against virtually any protein target. The generation of bicyclic peptides that advanced into clinical development involves the reaction of three cysteines in a peptide to a C3-symmetric alkylating agent. In phage display, this chemical modification transforms a pool of conformationally flexible peptides into a library of structurally unique protein mimetics that are able to bind traditionally challenging protein surfaces like those with limited structural definition. In recent years, a new class of bicyclic peptides has emerged using a single atom—bismuth—in place of C3-symmetric organic scaffolds, thus expanding into an unexplored chemical space at the intersection of inorganic chemistry and biology. This mini-review aims to reflect on the discovery, evolution and potential future applications of bismuth bicycle molecules.
Making Blank Faces Expressive: Chemical Approaches to the Modification of Chemically Inert Peptides

As an alternative to the conventional approach, which combines amino acid monomers in a one-by-one fashion to peptide derivatives, the chemical modification of existing peptides has attracted significant attention in recent years. However, such approaches generally target the reactive functional groups in cysteine and lysine residues, particularly in the case of larger peptides, and the modification of peptides that do not feature these functionalities is more difficult. This review focuses on recent attempts to achieve such more ambitious chemical modifications.
ABSTRACT
Very minor structural modifications in peptides can result in significant changes in their function. To obtain analogues with such small structural but big functional differences compared to the original peptides, amino acid monomers are usually combined one-by-one from scratch, which is a simple and reliable but painfully laborious strategy. One alternative and very fascinating approach is the chemical modification of existing peptides, which allows for the rapid production of derivatives with fewer synthetic steps. However, such approaches generally target the reactive functional groups in cysteine and lysine residues, particularly in the case of larger peptides, and the modification of peptides that do not feature these functionalities is more difficult. Nevertheless, chemists have also been exploring methods that can be applied even to such chemically inert peptides based on recent advances in C–H activation and hydrogen atom transfer (HAT) chemistry. If successful, these strategies would represent a breakthrough in terms of obtaining unusual peptide structures in a time- and cost-effective manner. This review focuses on recent attempts to achieve such ambitious chemical modifications, albeit that these are currently limited to relatively small peptides.
Identification of Human Bone Morphogenetic Protein‐2 Elbow Epitope as Its Coreceptor Recognition Site and Design of Elbow‐Derived Hairpin Peptides to Specifically Target the Coreceptor

An elbow epitope is identified as the specific binding site of BMP2 coreceptor, in addition to the previously reported wrist and knuckle epitopes serving as the recognition sites of BMP2 type-I and type-II receptors, respectively. Several bioactive peptides are successfully derived from the elbow epitope segment, which exhibit good binding potency to specifically target BMP2 coreceptor.
ABSTRACT
Human bone morphogenetic protein-2 (BMP2) is an effective osteoinductor in bone formation and growth via binding to its cognate receptors and coreceptors. Traditionally, the protein is identified to possess a conformational wrist epitope and a linear knuckle epitope that can be recognized by type I and type II receptors, respectively. In this study, we defined an additional epitope termed elbow as the specific binding site of its coreceptor, which is concentrated into a small helical hairpin region of BMP2 monomer 1 and partially overlaps with wrist but is far away from knuckle. A linear lEEP peptide is derived from the elbow epitope, which, however, is intrinsically disordered and cannot be maintained in native helical hairpin conformation due to lack of BMP2 protein context support, thus only exhibiting a low affinity to coreceptor. Disulfide stapling strategy is then used to stabilize the helical hairpin conformation of linear lEEP, which results in its two cyclic counterparts. The stapling is demonstrated to effectively improve peptide affinity and also exhibits a high selection for coreceptor over type-I receptor. The cyclic peptides are found to maintain a well-ordered, native-like conformation, which can be readily recognized by coreceptor through a conformational selection mechanism.
An Effective Method for Isolating Ergothioneine Analogues for Use in Peptide Synthesis: 2‐Thiohistidine and Fmoc‐2‐Thiohistidine

We present a simple method for isolating the novel amino acid 2-thiohistidine and its Fmoc-derivative through the addition of sodium chloride to the isolation step. The introduction of sodium chloride to this step greatly improves the yield and purity of the product. The metal binding utility of this amino acid should be further explored.
ABSTRACT
Here, we report on an improved synthesis of the novel amino acid 2-thiohistidine and its Fmoc-derivative through the addition of sodium chloride to the isolation steps of each synthetic procedure. These compounds must be synthesized in either water or a mixture of water/organic solvent and have proven difficult to isolate by traditional methods. We serendipitously discovered that the addition of sodium chloride caused each compound to precipitate from the solution during the workup. Since 2-thiohistidine is the amino acid analogue of the natural product ergothioneine, we hypothesize that sodium cations mimic the binding of other biologically relevant metal cations to the thioimidazole moiety. Ergothioneine is known to bind Cu+, Zn2+, and Hg2+, and it may bind other metals. Our fortuitous discovery is not only useful for the isolation and purification of 2-thiohistidine and its derivatives; peptides that contain this unique amino acid may be designed to take advantage of binding to Group I cations and other metals.
Turn‐Induction in Peptides Incorporating Novel Cyrene‐Derived α,α‐Disubstituted Amino Acid

Novel chiral cyrene-derived Cyr residues were incorporated into peptides. The α,α-disubstituted amino acid residue was introduced using the Ugi reaction. X-ray crystallographic analysis of R– and S-Cyr residues indicated preference to adopt left- and right-handed α-helical conformations, respectively. Propensity to adopt positions in β- and γ-turn hydrogen-bonded conformers was also indicated in the solid state and by NMR studies in solution, highlighting potential in peptide design and conformational control.
ABSTRACT
Cyrene-derived α,α-disubstituted amino acids (H-Cyr-OH) are introduced into peptides using the multiple component Ugi reaction. Conformational analysis of the Cyr monomer and dimer was performed by X-ray crystallography and NMR spectroscopy. In the solid state, the backbone torsion angle values of (4R)- and (4S)-Cyr were respectively characteristic of left- and right-handed α-helical conformers; however, the dimer appeared to adopt a 10-membered hydrogen bond in a distorted type III β-turn. In solution, solvent shielded amide NH hydrogens suggested that the Cyr residue could adopt the central position of γ-turn conformations. With conformational properties indicative of α-, β-, and γ-turns, the Cyr residue offers interesting potential as a novel chiral α,α-disubstituted amino acid for exploring chemical space.
A Novel Cell‐Penetrating Lipopeptide for DNA Binding, Self‐Assembly, and Delivery Into HeLa and Pluripotent Stem Cells

A lipopeptide is designed that contains an epitope from simian virus T-antigen (SV40T, PKKKRKV) conjugated to an N-terminal palmitoyl (C16-) moiety, which acts as a cell-penetrating lipopeptide, with additional aggregation propensity conferred by the lipid chain. The lipopeptide enables DNA delivery into the cytoplasm and nucleus.
ABSTRACT
A lipopeptide is designed that contains an epitope from simian virus T-antigen (SV40T, PKKKRKV) conjugated to an N-terminal palmitoyl (C16-) moiety, with the aim to act as an effective cell-penetrating lipopeptide, with additional aggregation propensity conferred by the lipid chain. A combination of cryo-TEM and small-angle X-ray scattering (SAXS) is used to show that the lipopeptide forms micelles, but mixtures with DNA lead to formation of fractal cluster-like co-assemblies due to intercalation of the DNA and peptide. Spectroscopic studies using fluorescence and circular dichroism (along with fiber X-ray diffraction) show that the peptide interacts with DNA and inserts into the groove. Confocal microscopy along with flow cytometry confirms delivery of DNA into both HeLa and mouse embryonic stem cells (mESCs) in pluripotent state, and the system shows excellent cytocompatibility as confirmed by MTT assays. Our data indicate that the lipopeptide may outperform the DNA transfection agent lipofectamine in DNA delivery into these stem cells and it enables DNA delivery into the cytoplasm and nucleus.
Issue Information
No abstract is available for this article.
Emerging Therapies in Metabolic Health: A Comprehensive Review of GLP‐1, GIP, and Glucagon Agonists

This review talks about the development of incretin agonists (glucagon-like peptide-1 [GLP-1], GIP [glucose-dependent insulinotropic polypeptide], glucagon), oral formulation development approaches, and advanced drug delivery platforms. It provides details about clinical trials on incretin agonists, focussing on efficacy, safety, and innovations in clinical research.
ABSTRACT
Incretin-based therapies have become central to type 2 diabetes (T2D) management, offering benefits beyond glycemic control, including weight reduction, cardiovascular protection, and emerging roles in renal and neurological health. This review addresses the question: What are the recent advances in GLP-1, GIP, and glucagon receptor agonists, and how do formulation strategies overcome biopharmaceutical challenges? We systematically analyzed late-stage clinical trials and formulation approaches for peptide-based therapies. The review focuses on therapeutic efficacy, structural and physicochemical properties influencing absorption, distribution, and metabolic stability, and strategies to mitigate degradation pathways such as enzymatic hydrolysis and peptide aggregation. Additionally, innovative delivery systems such as oral peptide formulations and long-acting injectables demonstrate promise in addressing inherent challenges of peptide drug delivery. By integrating clinical outcomes with mechanistic and formulation insights, this review highlights the evolving landscape of incretin-based therapies and underscores innovative solutions for peptide stabilization and delivery. These findings provide a forward-looking perspective for clinicians, researchers, and pharmaceutical scientists engaged in T2D management and drug development.
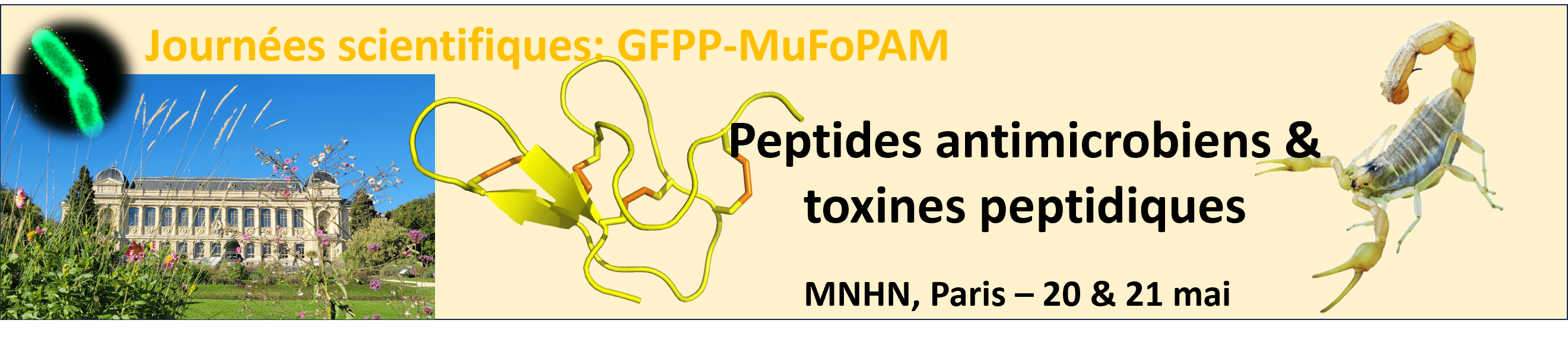
PAMTOX 2026
Les journées scientifiques « peptides antimicrobiens et toxines peptidiques » (PAMTOX) auront lieu les 20 et 21 mai 2026 au Muséum National d’Histoire Naturelle (MNHN, Paris). Ces journées sont co-organisées par le réseau Multifonction des Peptides AntiMicrobiens (MuFoPAM, https://www.mufopam.fr/) et le Groupe Français des Peptides et Protéines (GFPP, www.gfpp.fr). Les inscriptions sont en ligne, à …
Continue reading “PAMTOX 2026”
L’article PAMTOX 2026 est apparu en premier sur Groupe Français des Peptides et des Protéines.
PAMTOX 2026 meeting
The scientific meeting “Antimicrobial Peptides and Peptide Toxins” (PAMTOX) will take place on May 20–21, 2026 at the National Museum of Natural History (MNHN, Paris). The meeting is co-organized by the Multifunctional Antimicrobial Peptides Network (MuFoPAM) and the French Group for Peptides and Proteins (GFPP).
Registration is available online at the following address:
Antimicrobial peptides (AMPs) and peptide toxins are two major classes of bioactive peptides involved in defense or attack functions, depending on the organism. They share many points of convergence, both conceptually (chemical ecology, structures, activities, molecular-scale mechanisms) and in their methodological approaches (production techniques, integrative “omics” approaches, biophysical methods, etc.). AMPs and toxins offer strong potential in bioengineering and represent a valuable source of inspiration for the development of new therapeutic strategies.
The PAMTOX meeting will provide an opportunity to review these different aspects and to promote exchanges between scientific communities in a multidisciplinary context. It will bring together up to 100 participants, including biologists and chemists from both the public and private sectors.
The following themes will be addressed:
-
Diversity and ecological roles of antimicrobial peptides and toxins
-
Structures and mechanisms of action
-
Design and production through chemical synthesis and bioengineering
-
Applications and valorization
A significant part of the program will be devoted to presentations by early-career researchers and established scientists. Approximately 15 oral presentations will be selected from submitted abstracts, and space will be reserved for poster presentations.
 |
 |
 |
 |
 |
 |
Issue Information
No abstract is available for this article.
On‐Resin Pictet–Spengler Cyclization for 1,2,3,4‐Tetrahydro‐β‐carboline‐3‐carboxylate (Tcc) Peptide Synthesis With Acid‐Labile Functional Tolerance

The constrained tryptophan (Trp) analog 1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid (Tcc) is used to restrict peptide conformation in structure–activity relationship studies. Although Pictet–Spengler cyclization of Trp and aldehydes with mineral acid gives Tcc analogs, such harsh conditions do not tolerate acid-labile functionality. To enable broader use of Tcc residues, mild conditions are now reported for the on-resin Pictet–Spengler cyclisation in the presence of acid-labile side chain protection and linker strategies.
ABSTRACT
The constrained tryptophan (Trp) analog 1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid (Tcc) is used to restrict peptide conformation in structure–activity relationship studies. Although Pictet–Spengler cyclization of Trp and aldehydes with mineral acid gives Tcc analogs, such harsh conditions do not tolerate acid-labile functionality. To enable broader use of Tcc residues, mild conditions are now reported for the on-resin Pictet–Spengler cyclisation in the presence of acid-labile side chain protection and linker strategies.
GFPP26
Le prochain congrès du GFPP aura lieu à l’Île d’Oléron du 13 au 17 juin 2027
L’article GFPP26 est apparu en premier sur Groupe Français des Peptides et des Protéines.
GFPP26
Le prochain congrès du GFPP aura lieu à l’Île d’Oléron du 13 au 17 juin 2027
L’article GFPP26 est apparu en premier sur Groupe Français des Peptides et des Protéines.
GFPP26
Le prochain congrès du GFPP aura lieu à l’Île d’Oléron du 13 au 17 juin 2027
L’article GFPP26 est apparu en premier sur Groupe Français des Peptides et des Protéines.
Antiproliferative Activity of the New Analogues of Acridine/Acridone With Oligopeptide Derivatives and Molecular Docking With EGFR and Src Kinase

New functionalized analogues of 1-nitroacridine/4-methyl-1-nitroacridine and 4-nitroacridone connected to tuftsin/retro-tuftsin derivatives as a potential anticancer therapeutics.
ABSTRACT
The aim of our work was to analyze new functionalized analogues of 1-nitroacridine/4-methyl-1-nitroacridine and 4-nitroacridone connected to tuftsin/retro-tuftsin derivatives as a potential anticancer therapeutics. Using the MTT assay, we have evaluated the cytotoxic activity of synthesized analogues against the Jurkat cell line and healthy donor–isolated peripheral blood mononuclear cells (PBMCs). Jurkat cells were susceptible to several of the acridine derivatives, whereas PBMCs were sensitive to two compounds only. 4-Methyl-1-nitro-acridine analogue, classified as compound 28, has exhibited the greatest anticancer potential with 4-fold higher cytotoxicity toward Jurkat cells and 12-fold lower cytotoxicity against PBMCs when compared with control therapeutics. Subsequent molecular docking analysis suggested that binding to epidermal growth factor receptor (EGFR) and proto-oncogene tyrosine-protein kinase Src molecules could be, at least partially, responsible for observed biological effects of created acridine/acridone analogues.
Correction to “Discovery of Antioxidant Peptide Against Keap1 From Silkworm Protein Based on In Silico Study and In Vitro Antioxidant Activity Detection”
Journal of Peptide Science, Volume 32, Issue 1, January 2026.
Side Reaction Analysis in Solid‐Phase Peptide Synthesis: A Case Study in the Glu–Asp–Tyr Motif

In the Fmoc-SPPS of the Glu-Asp-Tyr motifs, Asi, Agl, and Pgl by-products are formed, amplified by microwave assistance. Their levels depend on the sequence, protecting groups, deprotection conditions, and the conformation/steric hindrance of the growing chain. Optimal minimization: 6% piperazine with 0.1 M oxime.
ABSTRACT
Peptides are important tools in biological, medical, and pharmaceutical research. Solid-phase peptide synthesis (SPPS) is the primary method for their preparation in high yields and purity. However, SPPS often encounters difficulties due to the occurrence of side reactions, which can generate by-products that are difficult to remove. Side reactions can occur under both basic and acidic conditions, resulting in reduced yields, costly purification processes, and sometimes potential inaccessibility of the target peptides. By-products include the formation of aspartimide, glutarimide, and pyroglutamic acid. The problem of unwanted intramolecular cyclizations is well known and unavoidable, in some cases minimized, as it depends on the intrinsic thermodynamics of the final structures. In this context, we report a systematic analysis of the side reactions that occur during SPPS in peptides containing the Glu-Asp-Tyr motif. This study is the first to investigate the formation of three different by-products within the same peptide sequence and to provide valuable insights into the experimental conditions that favor one reaction over the others. The study aims to identify the optimal experimental conditions to mitigate these by-products’ formation. Our results highlight that the steric and conformational effects of the growing protected peptide chain play a role that is often overlooked or misinterpreted.
Statuts du GFPP
ASSOCIATION GROUPE FRANÇAIS DES PEPTIDES ET DES PROTEINES STATUTS MIS A JOUR EN DATE DU 24/06/2025 ARTICLE 1. DÉNOMINATION Il a été fondé en 1992 une association régie par la loi du 1er juillet 1901 et le décret du 16 août 1901, sous la dénomination de GROUPE FRANÇAIS DES PEPTIDES ET DES PROTEINES (GFPP). …
Continue reading “Statuts du GFPP”
L’article Statuts du GFPP est apparu en premier sur Groupe Français des Peptides et des Protéines.
Rational Design, Synthesis, and Aphicidal Activity of Novel Insect Short Neuropeptide Analogs as Potential Aphid Control Agents

Ten novel sNPF analogs were designed and synthesized based on the structure of lead I-3 by molecular docking and peptidomimetic methods. Compound A-1 exhibits better binding affinity with the receptor, possesses excellent aphidicidal activity, and shows low toxicity to bees, making it a promising candidate for environmentally friendly insecticides.
ABSTRACT
Short neuropeptide F (sNPF) is a peptide unique to insects, characterized by a C-terminal phenylalanine and a conserved RLRFa motif, and plays key roles in controlling feeding behavior, growth, circadian rhythms, and water–salt homeostasis. We previously identified an sNPF analog, I-3, with aphidicidal activity. In this study, 10 sNPF analogs with aromatic or nonaromatic modifications at the N-terminus were designed based on I-3, using molecular docking and peptidomimetic strategies to investigate the role of N-terminal residues. Aphicidal activity showed that A-1 has stronger activity than I-3 and pymetrozine. Structure–activity analysis indicated that a benzene ring with electronegative and lipophilic groups at the N-terminus is key for aphicidal activity. Molecular docking and molecular dynamics simulations showed A-1 binds more stably to the receptor than I-3. Toxicity tests on honeybees (Apis mellifera) confirm that compound A-1, which exhibits strong aphidicidal activity, is safe for nontarget organisms. Additionally, Admetsar3 evaluations indicate low toxicity risks for all compounds. Therefore, A-1 represents a promising, selective, and eco-friendly insecticide for controlling pea aphids, and this study validates the feasibility of developing novel green pesticides based on sNPF.
Issue Information
No abstract is available for this article.
In Silico Peptide Design: Methods, Resources, and Role of AI

This review explores various computational methods based on structure and ligand-based approaches along with advanced AI models in transforming peptide design. We also discuss the impact of peptide databases in accelerating peptide research.
ABSTRACT
Peptides play essential roles in biological systems and serve as key agents in therapeutics, biomaterials, and drug delivery. Despite their broad utility, peptide design is limited by rapid degradation, low oral bioavailability, and the inefficiency of conventional synthesis and screening methods. This review provides a comprehensive overview of computational approaches that have emerged as effective alternatives, enabling the exploration of a large chemical space and the virtual screening of thousands of peptides. We detail the critical role of specialized peptide databases, computational tools, and advanced methodologies, including structure-based design, molecular dynamics (MD) simulations, and ligand-based approaches. A particular focus is placed on the transformative impact of machine learning (ML), deep learning (DL), and generative AI models, which are accelerating the discovery of novel peptides. While these methods offer promising solutions, we also address key challenges like data inconsistency, model interpretability, and the need for better forcefields. By highlighting these advancements and limitations, this review aims to provide a roadmap for leveraging computational design in peptide research.
Issue Information
No abstract is available for this article.
Estimation of Minimum Diversity to Be Prepared in Random Peptide Libraries for Target‐Binding Peptide Discovery

The theoretical diversity of peptide libraries is astronomical. We define a practical lower bound as Y ≥ 1/(s × a × ∏k=1mfk$$ {prod}_{k=1}^m{f}_k $$). A literature survey estimates that a diversity of ≈1.6 × 105 (m ≈ 4, a ≈ 1, s ≈ 1) is required to include one specific binder.
ABSTRACT
Molecular library display systems utilizing phage, bacteria, and genetic material are powerful tools for identifying target-specific peptides. Libraries of sufficient diversity are required to isolate target-specific peptides. Although the evolution of molecular display techniques—from phage display to mRNA display—has substantially expanded achievable library diversities, the minimum diversity required to reliably isolate target-specific peptides remains unclear. Here, we propose a straightforward equation to estimate the minimum diversity (Y). Y is defined by three experimentally accessible parameters: (i) the number of important amino acids (m, consensus motif residues whose substitution markedly reduces binding), (ii) the number of independent binding sites (s) on the target molecule and (iii) the arrangement factor (a) that counts the possible positional permutations of the motif within the random region. By analyzing 35 target-specific peptides reported previously, we found that the average value of m was ≈4, whereas a and s were most frequently ≈1. These representative values imply a practical lower-bound benchmark of diversity, Y ≥ 1.6 × 105 for random peptide libraries in screening. Our findings will aid researchers in rationalizing the design and construction of peptide libraries, facilitating efficient identification of high-affinity, target-specific peptides.
Metal Ion–Induced Cross‐Linking in Mucin‐Inspired Peptide Hydrogels

Short fiber-forming peptides were modified by histidine substitution and mannose or galactose attachment and combined with different metal ions. Although no generalizable trends were observed, Lys to His substitution at position 25 with galactose in the presence of copper produced a species with highly favorable viscoelastic properties.
ABSTRACT
Mucus is the biological hydrogel that lines the mucosal surfaces of mammals and acts as a protective barrier. Its main proteinaceous component is mucin, the high molecular weight, degree of glycosylation, and hardly uniquely defined nature of which hamper precise structures/property investigations based on biological samples. In contrast, chemically precisely defined peptide model systems inspired by such natural glycoproteins represent synthetically readily obtainable tools with excellent properties for both fundamental research and biomedical applications. Herein, we report the design and characterization of a library of histidine- and monosaccharide-containing coiled coil peptides that form hydrogels to different degrees in the presence of divalent metal ions Cu2+, Zn2+, Ca2+, and Fe2+. Using rheology, circular dichroism, and transmission electron microscopy, we determined the viscoelastic properties and global structures of these glycopeptide materials. This study reflects the interplay between glycan identity, histidine position, and divalent metal ion on the mechanical strength of these hydrogels.
Antimicrobial Peptide Moricin Inhibits Streptococcus pneumoniae Growth Through Membrane Disruption: Insights From In Silico and In Vitro Studies

In vitro dose–response tests reveal that moricin peptide retards the growth of Streptococcus pneumoniae. Molecular docking and MD simulation tests show that moricin binds strongly to Pneumococcal Surface Adhesin A (PsaA). ADMET profile shows that moricin can be safely absorbed, has minimal toxicity and could be used as a treatment. Moricin disrupts membrane potential and causes membrane leakage of bacteria. It is biocompatible, with no significant toxicity to BV2 microglial cells.
ABSTRACT
The rise of multidrug-resistant Streptococcus pneumoniae poses a major public health threat, necessitating novel therapeutic targets. Pneumococcal Surface Adhesin A (PsaA), a conserved surface lipoprotein, plays a key role in manganese acquisition, colonization and virulence. Immunization with PsaA elicits protective immunity, while fragment-based drug design has identified inhibitors disrupting its function. PsaA emerges as a promising molecular target for innovative therapeutic strategies against pneumococcal diseases. Antimicrobial peptides (AMPs) are crucial components of the innate immune system, providing a potent defence mechanism against a broad spectrum of pathogens. Moricin, an AMP initially identified in Bombyx mori, exhibits robust antimicrobial activity against Gram-positive bacteria. This study explores the inhibitory effects of moricin on S. pneumoniae, a significant human pathogen responsible for severe infections such as pneumonia, meningitis and sepsis. In silico analyses, including molecular docking and molecular dynamics simulations, revealed a strong interaction between moricin and the PsaA. In vitro studies corroborated the computational findings, demonstrating a dose-dependent inhibition of S. pneumoniae growth. Moricin induced bacterial membrane disruption, evidenced by increased membrane permeability, release of intracellular contents and altered membrane potential, highlighting the bactericidal mode of action. Furthermore, time-kill kinetics revealed rapid bacterial eradication, underscoring moricin’s efficacy. Additionally, toxicity assays on the macrophage BV2 cell line demonstrated that moricin caused no significant structural or organelle damage, emphasizing its biocompatibility and safety. The integration of in silico and in vitro approaches provides comprehensive mechanistic insights into moricin’s antimicrobial action and establishes its potential as a safe and effective therapeutic agent against S. pneumoniae.
Peptide Tags for Site‐Selective Nonenzymatic Covalent Modification of Proteins

Highly selective chemical modification of peptides. This review describes peptide tags that direct covalent chemical modification of proteins.
ABSTRACT
Bioconjugation chemistry is an important tool for studying proteins, developing pharmaceutical agents, and for many other applications. Conventional methods for protein functionalization rely on chemoselective reactions but often have poor regioselectivity. Peptide tags facilitating site-selective chemical covalent modification of proteins are of great value in the synthesis of protein conjugates. Ideally, a protein would only have to be minimally mutated prior to chemical modification to avoid interfering with native protein folding, trafficking, and function. This short review summarizes the advances in the developments and applications of peptide tags for covalent modifications that proceed without enzymatic assistance.
Plant Hormone Cytokinin as Aggregation Modulator of Gelsolin Amyloidosis

Gelsolin peptides form amyloids, and aggregation is accelerated by a single point mutation. The gelsolin amyloids can be inhibited using plant hormones.
ABSTRACT
Amyloidosis, a self-assembly of proteins or peptides, is associated with numerous degenerative diseases, such as gelsolin amyloidosis, which remain without a cure. Gelsolin protein is an actin-binding protein, but when aggregated in a diseased state, it is a potential drug target. Specifically, gelsolin mutations, N184K and D187Y, have been linked to renal amyloidosis and systemic progressive deposition of amyloids, respectively. Understanding how such mutations mitigate gelsolin aggregation and how this process can be prevented through small molecule inhibitors is of interest. Herein, we explored the efficacies of plant-based naturally occurring cytokinin (CK) molecules as aggregation modulators in vitro. Using various biophysical methods, such as spectroscopy and microscopy, the aggregation of wild-type gelsolin peptide 184NNGDCFILDL193 and its mutants (N184K, D187Y) was investigated. The mutations significantly promoted aggregation, which is of biological significance. The CK trans-zeatin (tZ) was a more effective disaggregation promoter compared with kinetin (Kin). The experimentally determined IC50 values were in the 9–20 μM range. The mode of inhibition was identified as direct non-covalent complexation between the CK and the peptides by using mass spectrometry and molecular docking studies. Data show that CKs are promising amyloid modulators, which can be easily translatable to other amyloid systems.
Discovery of a Thrombin‐Targeting Anticoagulant Peptide From Whitmania pigra via a “Computation‐Guided Experimentation” Strategy

A peptide library from nonbloodsucking leech Whitmania pigra was established. Through molecular docking and dynamics (MD) simulations, a thrombin-targeting PEPWP (LRELEDALEQER) was screened out, showing significant in vitro/vivo anticoagulant activity. Thrombin’s Arg233 and Arg101 in Exosite II were revealed as the key electrostatic interaction site, distinguishing its mechanism from hirudin.
ABSTRACT
Targeting thrombin to screen safe thrombin inhibitors from natural plants and animals is a critical direction in anticoagulant drug development. This study aimed to screen thrombin inhibitors from the nonbloodsucking leech Whitmania pigra (WP) and elucidate the mechanism of anticoagulation through a “computation-guided experimentation” strategy. A peptide library was constructed from WP hydrolysates, and virtual screening was performed using molecular docking and dynamics simulations. A novel thrombin-targeting anticoagulant peptide PEPWP (LRELEDALEQER) was screened out from the peptide library and validated through in vitro/in vivo experiments. PEPWP significantly prolonged thrombin time (TT) and prothrombin time (PT) in a dose-dependent manner in vitro, indicating its role in the common and extrinsic coagulation pathways. Surface plasmon resonance (SPR) analysis then confirmed strong thrombin binding (K
d
= 7.242 × 10−6 mol/L). Furthermore, PEPWP prolonged TT while reducing blood viscosity in acute blood stasis rats. Finally, structural analysis revealed that PEPWP bound to Exosite II of thrombin. Arg233 and Arg101 were the key residues for the binding. In conclusion, PEPWP exhibited good anticoagulant activity and significant application potential.
Amphipathic Octenyl‐Alanine Modified Peptides Mediate Effective siRNA Delivery

This study demonstrates that novel octenyl-alanine–modified hPep peptides efficiently encapsulate siRNA into well-defined nanoparticles. Our findings highlight the potential of these hPep/siRNA nanoparticles to induce RNA interference, effectively silencing therapeutically important CD45 expression.
ABSTRACT
The development of therapeutic small interfering RNAs (siRNAs) has lately gained significant momentum due to their ability to silence genes in a highly specific manner. The main obstacle withholding the wider translation of siRNA-based drug modalities is their limited half-life and poor bioavailability, especially in extra-hepatic tissues. Consequently, various drug delivery systems (DDSs) have been developed to improve the delivery of siRNAs, including short delivery peptides called cell-penetrating peptides (CPPs). In this study, we explore the potential of using alkenyl-alanine modifications to enhance the siRNA delivery efficacy with CPPs. We demonstrate on hPep peptides that incorporation of alkenyl-alanines enhances the encapsulation of siRNAs into stable nanoparticles and contributes to increased cellular uptake. Furthermore, we demonstrate that the lead peptide, hPep3, induces effective RNAi-mediated gene silencing in a reporter cell model as well as on the disease-implicated endogenous CD45 gene target. The biodistribution studies in mice show that the alkenyl-alanines are systemically well tolerated, and employing such modifications in the peptide backbone improves siRNA delivery in several tissues, including extra-hepatic sites. As demonstrated on hPep peptides, alkenyl-alanines offer a simple yet robust way to enhance the delivery efficacy of CPPs and have the potential to advance siRNA therapeutics beyond the liver targets.
Leveraging the Therapeutic Potential of Natural Peptide Panurgines: Hydrocarbon Stapling Strategy Enhances Their Efficacy Against Breast Cancer

Stapled Panurgines peptides were developed to enhance stability and permeability. PNG-5 showed improved helicity, protease resistance, and anti-breast cancer activity, highlighting the potential of hydrocarbon stapling for creating effective peptide-based therapeutics.
ABSTRACT
The development of novel candidate molecules for breast cancer treatment holds significant clinical value. Panurgines (PNG), derived from the venom of the wild bee Panurgus calcaratus, are particularly noteworthy for their anti-breast cancer activity and antibacterial properties. However, linear peptides are often hindered by poor stability and limited cell membrane permeability, making them highly susceptible to protease degradation. To tackle this challenge, the current study focused on synthesizing a range of stapled Panurgines peptides through hydrocarbon stapling modifications, followed by a comprehensive evaluation of their chemical and biological properties. Remarkably, PNG-5 demonstrated notable improvements in helicity, cell membrane permeability, proteolytic stability, and antitumor activity. This study examines how the hydrocarbon stapling technique significantly affects the secondary structure, hydrolase stability, and biological activity of PNG, revealing its potential as a transformative and powerful anti-breast cancer therapeutic. These findings lay a strong foundation for the development of innovative and highly effective peptide-based anti-tumor drugs.
DiPTH‐Cystine and PTH‐Cysteine in Disulfide Bond Analysis Using Automated Edman Degradation

The mystery of PTH-cysteine and diPTH-cystine in the analysis of disulfide bonds is solved. Both derivatives were synthesized and applied to Edman sequencing. A time-efficient combined workflow applying Edman and MS/MS sequencing for a comprehensive analysis of the complex disulfide connectivity in peptides and proteins was developed.
ABSTRACT
The annotation of disulfide bridges in peptides and proteins can be an elaborate process and requires careful revision of multiple data sets to avoid wrong assignment in the structural analysis. Herein, we provide additional support to elucidate the cysteine connectivity by re-implementation of Edman sequencing for the analysis of this specific structural feature. By synthesizing diPTH-cystine and PTH-cysteine for comparison, we were able to identify the respective derivative during Edman sequencing when a disulfide bond is detected in a peptide. Application of Edman sequencing to selected peptides with two or three disulfide bridges provides further insight into the differentiation of cysteines that form a disulfide bridge for both half-cystines in the same cycle and in separated cycles. A combined approach for the implementation of automated Edman sequencing in the process of disulfide bond assignment is described to alleviate structural elucidation in the future analysis of cysteine-rich peptides and proteins.
Issue Information
No abstract is available for this article.
Silica‐Assisted Solid‐Phase Peptide Synthesis (SiPPS)

The use of a non-swelling resin for solid-phase peptide synthesis allows the reduction of solvent consumption by up to 50%.
ABSTRACT
Non-swelling silica-based resin was used for peptide synthesis. The strategy used is similar to that of solid-phase peptide synthesis (SPPS), referred to as silica-assisted solid-phase peptide synthesis (SiPPS). A 2-h coupling seemed to favor the coupling compared to that of 1-h standard coupling. The use of this non-swelling resin allows a 50% reduction in the consumption of solvents. The strategy was well demonstrated for the synthesis of H-YSSFL-NH2, linear oxytocin, angiotensin II, and afamelanotide using a silica-based support (Fmoc-Rink amide SiliaBond manufactured from SiliCycle Inc.). The peptides were found to have more than acceptable purity, although there was a loss in overall yields.
Discovery of Antioxidant Peptide Against Keap1 From Silkworm Protein Based on In Silico Study and In Vitro Antioxidant Activity Detection

A novel antioxidant peptide WQK was identified from Silkworm pupa by virtual digestion, ADMET prediction, and molecular docking. WQK can reduce ABTS·+ and ferric-tripyridyltriazine, in vitro. Besides, WQK can promote the expression of antioxidant genes and eliminate reactive oxygen species in oxalic acid-treated human proximal tubular epithelial cells (HK-2).
ABSTRACT
Nrf2-Keap1 is an important defense system against oxidative stress damage and enhances the body’s antioxidant capacity. Targeting the Keap1-Nrf2 signaling pathway and activating Nrf2 has become an effective strategy for treating oxidative stress and related diseases. In this study, virtual digestion, ADMET prediction, and molecular docking were used to screen antioxidant peptide from silkworm pupa protein. Then, a novel tripeptide (WQK) was identified, exhibiting good water solubility, nontoxicity, high intestinal absorption, and the ability to cross the blood–brain barrier. Molecular docking showed that WQK established six H-bond interactions with some key sites of Keap1 (Arg380, Arg415, Gln530, Ser555, Ile416, and Leu365), which is similar to the identified antioxidant molecule 12e. Additionally, WQK has the function to reduce ABTS·+ and ferric-tripyridyltriazine (Fe3+-TPTZ) in vitro. Furthermore, WQK can promote the expression of antioxidant genes and eliminate reactive oxygen species (ROS) in oxalic acid-treated human proximal tubular epithelial cells (HK-2). The results suggested that WQK may activate the Keap1-Nrf2 signaling pathway by binding to Keap1 and activating the antioxidant system.
Interactions of Peptide Amphiphiles With Viruses and Cells Are Enabled by Amorphous Nanostructures

Amorphous nanostructures from polyunsaturated peptide amphiphiles such as eicosapentaenoyl-VVVAAAKKK-NH2 promote gene delivery by viral vectors.
ABSTRACT
Peptide amphiphiles can form fibrillar and amorphous structures. While fibrillar assemblies have previously been shown to enhance viral infectivity or retroviral transduction for gene delivery, we now elucidate the mechanism behind amorphous peptide amphiphiles that promote virus–cell interactions. Using electron microscopy, we reveal that amorphous fragments of polyunsaturated peptide amphiphiles allow for more VLPs to bind directly to the plasma membrane, explaining previously observed efficient viral entry and superior biodegradation compared to state-of-the-art adjuvants. We believe our work highlights the potential of unsaturated fatty acid peptide hybrid materials for clinical applications.
Assessment of Thermal and Photolytic Stress Effects on the Stability of Primary Structure of Synthetic Liraglutide Using LC‐HRMS/MS

Thermal and photolytic stability of liraglutide was studied, as it is essential to understand the effects of potential in-use mishandling and environmental stress conditions to understand its stability and degradation. Techniques, such as RP-HPLC and LC-HRMS (Orbitrap) were utilized to identify and characterize the generated impurities.
ABSTRACT
Liraglutide (LGT), a synthetic glucagon-like peptide-1 (GLP-1) analogue, is widely used in the treatment of Type 2 diabetes and obesity. Due to its peptide-based nature, it is prone to degradation, particularly under improper storage or handling conditions. Environmental stressors, such as heat and light, can lead to the formation of impurities that may compromise the quality and safety of the drug. In this study, we have investigated the impact of thermal and photolytic stress on LGT stability using reverse-phase liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS). This method enabled the separation, identification, and characterization of intact LGT and its degradation products (DPs). Ten impurities were identified, including four DPs that have not been reported so far. Further, MS/MS fragmentation analysis was employed to elucidate the sequences of LGT and its impurities, pinpointing the modifications and their respective sites. The formation mechanisms of these impurities were also proposed, providing critical insights into the degradation pathways. This study offers a comprehensive analytical approach for assessing the stability of peptide drugs, contributing to enhanced quality control and safer pharmaceutical formulations.
Modulating Antimicrobial Activity and Structure of the Peptide Esc(1‐21) via Site‐Specific Isopeptide Bond Formation

The introduction of an isopeptide bond at the lysine residues in the backbone of Esc(1-21) led to the generation of five analogs. Among these, Esc(1-21)ε20 showed the most promising features, having antimicrobial activity comparable with those of the parent peptide, a lower cytotoxicity, and a higher stability to proteolytic degradation.
ABSTRACT
Antimicrobial peptides (AMPs) represent valid alternatives to conventional antibiotics primarily due to their mechanism of action, which consists of cytoplasmic membrane disruption. However, their clinical application is often limited by cytotoxicity at high concentrations and low intrinsic biostability. To address these limitations, various biochemical approaches have been explored. In recent years, the frog-skin derived AMP Esc(1-21) has been extensively characterized for its potent antimicrobial activity, especially against Gram-negative bacteria, both in vitro and in vivo. In this study, we designed and synthesized novel Esc(1-21) analogs in which a single isopeptide bond was introduced in place of a conventional peptide bond at specific positions within the sequence. The resulting five analogs were evaluated for their (i) chemical and structural properties, (ii) resistance to proteolytic degradation, (iii) antimicrobial and antibiofilm activities, (iv) hemolytic and cytotoxic effects, and (v) ability to perturb bacterial cytoplasmic membranes. Among these, Esc(1-21)ε20 showed the most promising features, maintaining antimicrobial and antibiofilm activities comparable to those of the parent peptide while exhibiting lower cytotoxicity towards eukaryotic cells at higher concentrations and greater resistance to enzymatic degradation. These findings highlight Esc(1-21)ε20 as an attractive lead candidate for the development of new antibiotic therapeutics.
Modulation of Antimicrobial Activities of Aib‐Based Artificial Amphipathic α‐Helical Peptides by Incorporating Histidine Residues

Regarding the acquisition of antibacterial activity of BKBA-20 derivatives against Gram-negative bacteria by histidine incorporation, the suitable introduction position depends on pH.
ABSTRACT
Cationic antimicrobial peptides (CAMPs) exhibit potent antibacterial activity by disrupting bacterial membranes. We investigated the effect of histidine incorporation on BKBA-20, a designed amphiphilic helical peptide composed of alternating 2-aminoisobutyric acid (Aib) and lysine. Substitution at lysine sites (1a–1e series) reduced net charge and antimicrobial activity, though certain analogues (1c, 1d) demonstrated minimal antibacterial activity against Escherichia coli. In contrast, substitution at Aib sites (2a–2c series) preserved some extent of helical structure and improved activity under acidic conditions. Notably, substitutions at the terminal of the peptide were more effective at acidic pH, while the slightly medial side of the peptide favored activity at neutral pH. Hemolysis assays confirmed low cytotoxicity of the modified peptides. These results suggest histidine incorporation as a promising strategy to broaden the spectrum of CAMPs, particularly against Gram-negative bacteria, without increasing toxicity.
Peptide Synthesis Nomenclature
Journal of Peptide Science, Volume 31, Issue 9, September 2025.
Controversial Nomenclature in Peptide Synthesis: A Call for Clarity

The three main approaches commonly employed for peptide synthesis are solution synthesis or also called classical solution-phase peptide synthesis (CSPS), solid-phase peptide synthesis (SPPS), and liquid-phase peptide synthesis (LPPS).
ABSTRACT
Various approaches to make peptides have been adopted globally owing to their high demand. The three main approaches commonly used for this purpose are solution synthesis, also called classical solution-phase peptide synthesis (CSPS), solid-phase peptide synthesis (SPPS), and liquid-phase peptide synthesis (LPPS). Each method offers unique advantages: CSPS for scalability, SPPS for automation and efficiency, and LPPS for combining solution-phase simplicity with iterative synthesis using soluble tags.
Therapeutic Potential of Stylissatin A and Related Cyclic Peptides From Marine Sponges

Synthesis and biologicals activities of the natural cyclic peptide stylissatin A and its various analogues arereported. The findings suggest that specific amino acid residues such as Ile at position 2, Pro at 4 and 6, and Tyr at position 1 are key contributors to the peptide’s therapeutic potential.
ABSTRACT
Marine sponges are sessile invertebrates found in moderate, arctic, and tropical regions, serving as a valuable reservoir of bioactive compounds, particularly Pro-rich peptides. Among these, cyclic peptides have attracted significant interest due to their diverse therapeutic properties. One notable example is Stylissatin A (SA), a Pro-rich cyclic peptide reported from the marine sponge Stylissa massa. SA and its analogues have shown promising biological activities, including anti-inflammatory, anticancer, and anti-obesity effects. Despite the vast potential of marine-derived peptides, only a small number have progressed to the pharmaceutical market. Cyclic peptides like SA offer unique opportunities for molecular modifications and total synthesis, enabling the enhancement of potency, improvement of physicochemical properties, and optimization of synthetic yields. This review highlights the synthetic strategies developed for the total synthesis of SA, explores its structural features and related analogues, and discusses their therapeutic potential, underscoring the promise of SA-based scaffolds as novel peptide-based drug candidates.
Issue Information
No abstract is available for this article.